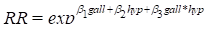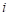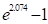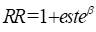Stratified models for the odds ratio – matched case control studies
This section contains examples that illustrate the use of
GMBO/PECAN to fit stratified models for the odds ratio. The primary focus
here is on the analysis of matched case control studies using conditional
logisticregression, but one can also fit stratified models using unconditional
logistic regression. The latter approach is useful, and is the EPICURE
default choice, for strata (case-contral sets) with large (more than 50 by
deafult) individuals.
The data used for most of the examples originated in a
matched case-control study of the effect of exogenous estrogens on the risk of
endometrial cancer among women in a retirement community near Los Angeles (Mack,
Pike et al. 1976). The study incldued 63 cases with 4 matched
controls per case. Matching was made on the basis of age (within one year),
vital status and community of residence at the time of diagnosis of the case,
age at entry into the retirement community, and marital status. Women who had a
hysterectomy prior to the time at which the case was diagnosed were not
considered as controls. The analysis of these data have been discussed in detail
by Breslow and Day (Breslow
and Day 1980).
The variables in this data set are
|
setno |
Matched set number |
|
cases |
Case-control indicator |
|
gall |
Gall bladder disease indicator |
|
hyp |
Hypertension indicator |
|
ob |
Obesity indicator |
|
est |
Estrogen usage indicator |
|
cdose |
Coded conjugate estrogen dose |
|
dura |
Estrogen usage duration in months |
|
non |
Non-estrogen drug usage indicator |
|
age |
Age at time of case diagnosis
|
The case-control indicator is coded 1 for cases and 0 for
controls. The other indicator variables are coded 1 for yes and 0 for no.
Conjugated estrogen dose is coded as 0 for none, 1 for -0.1 -0.299 mg/day, 2 for
0.3 -0.625 mg/day, and 3 for 0.6325 or more mg/day. Data on obesity, estrogen
dose, and estrogen usage duration were missing for some women. Missing values
for these variables are coded as 9, 9, and 99, respectively. The data are sorted
by set number, and within each set the case is the first.
Example
5.11 Matched Case-Control Data
with 1:1 Matching
For this example, we will reproduce some of the analyses
described in Section 7.3 of Breslow and Day (Breslow
and Day 1980). These analyses make use of 63 case-control pairs
consisting of the case and first control from each matched set in the data set.
Transformations will be used to create a variable that is used to select desired
records.
The commands used to read the input data (which are assumed to
be stored in a text file suitable for free-format input) and create some
additional variables needed for the analyses are
NAMES setno cases gall hyp ob est cdose dura non age
@
INPUT ../exdata/leimod.dat@
MISS cdose 9; ob 9; dura 99 @
CONS #cage = -1 @
TRAN IF cases == 1 THEN
#cage = age ;
ENDIF ;
cage = #cage ;
categ = 1 + (cage >= 65) + (cage >= 75) ;
cage70 = cage - 70 ; agegp = (age >= 70) ;
cest = (cdose > 0) ;
caseno = GL(5,1) ; pairs = caseno <= 2 ;
@
SAVE
@
The data are read from a file called LEIMOD.DAT. The MISS command is used to recode
missing values for dose, ob, and dura to the EPICURE missing value code. The constant
transformation is used to initialize the named constant (#cage) needed
for computation of the special age variable (cage) used in many of the
Breslow and Day analyses. This variable is defined to be the age of the case in
each matched set.
The GL function generates a sequence
of regular labels. In this case there are five labels and each label is repeated
one time, resulting in the sequence 1, 2, 3, 4, 5, 1 ,2, 3, 4, 5, 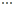 . Because of the way in which the data
are sorted, the value of caseno is 1 for the cases and 2 for the first
control for each case. The pairs variable is an indicator used to select records
for the initial analyses. The data are saved in a BSF file with the default
name, LEIMOD.BSF.
. Because of the way in which the data
are sorted, the value of caseno is 1 for the cases and 2 for the first
control for each case. The pairs variable is an indicator used to select records
for the initial analyses. The data are saved in a BSF file with the default
name, LEIMOD.BSF.
Since the analyses in this example will use only one control
per case, we select those cases with pairs equal to 1. This is accomplished with
the command
SELECT pairs == 1 @
Although it is not necessary for this example because the
default names are used, we explicitly specify the case-control indicator and
set-number variables using the commands. The COND
command instructs the program to use conditional likelihood.
CASES cases
STRATA setno@
COND @
We continue this example by reproducing some of the results
described in Table 7.3 of (Breslow
and Day 1980). The first model to be considered is a model with no
covariates, that is, a model in which none of the covariates have an effect on
the cancer risk. The NULL command is given after the
fit to designate this model as the null model for a subsequent likelihood ratio
test. This will create named constants called #_nuldv
and #_nulnp, which contain the deviance and degrees of
freedom, respectively, for this model. We also use constant transformations to
save another copy of the deviance for this model in a named constants called
#basdv for use in later computations. The commands to
fit this simple model and save the deviance are
FIT @
NULL
CONS #basdv = #_dv @
We will now compute score and likelihood ratio tests for an
effect of estrogen usage on the risk of endometrial cancer. The risk model used
here is the usual log-linear model

After computation of the score test, we fit the model and
compute the likelihood ratio test of the null hypothesis that estrogen usage has
no effect on the risk of endometrial cancer. In addition, we compute the Wald
confidence bounds (this also displays the relative risk estimate). This is done
using the following commands:
SCORE + est @
FIT @
LRT
CI @
The SCORE
command updates the model and computes the score statistic for the null
hypothesis that estrogens have no effect on the risk of endometrial cancer. The
FIT command is used to fit the updated model, and
CI causes 95 percent Wald bounds to be computed and
displayed. The LRT command instructs the program to
compute and display the likelihood ratio test. The output produced by these
commands is shown in Output
5.11.
The summary for the fitted model differs slightly from that for
an unstratifeid model. The model description and parameter summary table are
essentially identical. However, the deviance summary is different. In EPICURE
the “deviance” for conditional logistic regression models is defined as minus
two times the log-likelihood. This is identical to the goodness-of-fit ( ) statistic of Breslow and Day
(Breslow
and Day 1980). Defining the deviance in this way means that the
likelihood ratio statistic for the comparison of nested models can be computed
simply as the difference between the deviance values for the two fits with
degrees of freedom equal to the difference in the number of free parameters in
the model. The PECAN deviance summary also includes the number of free
parameters in the model, the number of risk sets used in the analysis, and the
number of
non-informative risk sets. Non-informative risk sets are those
risk sets in which the cases and controls have the same pattern for all
covariates in the model. In this model, there are 31 case-control pairs in which
both the case and control used or did not use estrogens and, thus, provide no
information for the estimation of the odds ratio non-informative.
) statistic of Breslow and Day
(Breslow
and Day 1980). Defining the deviance in this way means that the
likelihood ratio statistic for the comparison of nested models can be computed
simply as the difference between the deviance values for the two fits with
degrees of freedom equal to the difference in the number of free parameters in
the model. The PECAN deviance summary also includes the number of free
parameters in the model, the number of risk sets used in the analysis, and the
number of
non-informative risk sets. Non-informative risk sets are those
risk sets in which the cases and controls have the same pattern for all
covariates in the model. In this model, there are 31 case-control pairs in which
both the case and control used or did not use estrogens and, thus, provide no
information for the estimation of the odds ratio non-informative.
Output
5.11 Estimation and testing for an estrogen effect on endometrial
cancer risk-1:1 matching
FIT @
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Stratification on setno with 63 strata
Conditional likelihood used for 63 strata
Using pairs == 1
cases is used for cases
Records used 126
Deviance
87.3365 Free
parameters 0
Informative strata 63 (handled with the conditional
likelihood)
NULL
CONS #basdv = #_dv @
SCORE + est @
The score statistic is 21.125 with df = 1 (P <
0.001 )
FIT @
Iter Step Deviance
0 0 87.3365
1 0 64.2288
2 0 62.9341
3 0 62.8875
4 0 62.8874
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Stratification on setno with 63 strata
Conditional likelihood used for 63 strata
Using pairs == 1
cases is used for cases
Parameter Summary Table
#
Name
Estimate Std.Err. Test Stat. P value
-- ----------------------------
---------- --------- ---------- --------
Log-linear term 0
1
est......................
2.269 0.6065
3.741 < 0.001
Records used 126
Deviance
62.8874 Free
parameters 1
Informative strata 32 (handled with the conditional
likelihood)
Non-Informative strata 31
LRT
LR statistic
24.45 Degrees of
freedom 1
P value < 0.001
CI @
95%
Confidence Bounds
#
Name
Estimate Std. Err.
Lower Upper
-- ----------------------------
---------- --------- --------
--------
Log-linear term 0
1
est......................
2.269 0.6065
1.080 3.457
EXP(estimate)
9.667
1.834
2.945 31.73
The next set of commands are used to fit a set of nested models
for the effects of gallbladder disease, hypertension, and the joint effect of
having had both of these conditions on endometrial cancer risk. The models to be
used in these analyses are all contained in the following model:
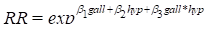
Before fitting the gallbladder model, we compute a score
statistic for the gallbladder disease effect. This could be done in a similar
way for the subsequent models. After the first fit, we use the LRT command to compute the likelihood ratio statistic and
then save the information about the current fit for use as the null model in the
next test. After the final test, we explicitly compute the two degree-of-freedom
likelihood ratio test comparing the full model to the model with no parameters
and use the CHISQ command to
evaluate the P-value for this test. If we had not given the NULL command after the first fit, the LRT command could have been used to compute this statistic
and the associated P-value. However, since the null model was changed, it is
necessary to compute the test statistic and its associated P-value explicitly.
This computation makes use of the #basdv constant saved earlier. The commands to
fit these models and carry out the tests of interest are
NOMODEL
FIT @
SCORE + gall @
FIT @ LRT
NULL
SCORE +hyp @
FIT @
LRT NULL
CONS #dvdif = #basdv - #_dv @
CHISQ 2 #dvdif @
FIT + gall*hyp @ LRT
This series of commands is generally straightforward; however,
you should note the use of the NOMODEL command to clear the
previous model. This command removes all parameters currently in the model.
(This could also be accomplished with the LOGL 0 @
command.) This is done here only because we wish to compute the score test. A
portion of the output produced by these commands is shown below.
Output 5.12 Output (partial) for estimation and
testing for the effects of gallbladder disease and hypertension on the risk of
endometrial cancer - 1:1 matching
NOMODEL
FIT @
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Stratification on setno with 63 strata
Conditional likelihood used for 63 strata
Using pairs == 1
cases is used for cases
Records used 126
Deviance
87.3365 Free
parameters 0
Informative strata 63 (handled with the conditional
likelihood)
SCORE + gall @
The score statistic is 3.55556 with df = 1 (P
= 0.0593)
FIT @
Iter Step Deviance
0 0 87.3365
1 0 83.6698
2 0 83.6536
3 0 83.6536
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Stratification on setno with 63 strata
Conditional likelihood used for 63 strata
Using pairs == 1
cases is used for cases
Parameter Summary Table
#
Name
Estimate Std.Err. Test Stat. P value
-- ----------------------------
---------- --------- ---------- --------
Log-linear term 0
1
gall.....................
0.9555 0.5262
1.816 0.0694
Records used 126
Deviance
83.6536 Free
parameters 1
Informative
strata 18 (handled with the conditional likelihood)
Non-Informative strata 45
LRT
LR statistic
3.683 Degrees of
freedom 1
P value 0.0550
NULL
SCORE +hyp @
The score statistic is 0.861336 with df = 1 (P
= 0.353)
FIT @
Iter Step Deviance
0 0 83.6536
1 0 82.7887
2 0 82.7878
3 0 82.7878
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Stratification on setno with 63 strata
Conditional likelihood used for 63 strata
Using pairs == 1
cases is used for cases
Parameter Summary Table
#
Name
Estimate Std.Err. Test Stat. P value
-- ----------------------------
---------- --------- ---------- --------
Log-linear term 0
1
gall.....................
0.9704 0.5307
1.828 0.0675
2
hyp......................
0.3481 0.377
0.9233 0.356
Records used 126
Deviance
82.7878 Free
parameters 2
Informative strata 39 (handled with the conditional
likelihood)
Non-Informative strata
24
LRT
LR statistic 0.8657
Degrees of freedom 1
P value 0.352
NULL
CONS #dvdif = #basdv - #_dv @
CHISQ 2 #dvdif @
Chi square = 4.5487 df = 2 P = 0.102864
FIT + gall*hyp @
Iter Step Deviance
0 0 82.7878
1 0 80.8714
2 0 80.8413
3 0 80.8413
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Stratification on setno with 63 strata
Conditional likelihood used for 63 strata
Using pairs == 1
cases is used for cases
Parameter Summary Table
#
Name
Estimate Std.Err. Test Stat. P value
-- ----------------------------
---------- --------- ---------- --------
Log-linear term 0
1
gall.....................
1.517
0.699
2.17 0.03
2
hyp......................
0.6270 0.4353
1.44 0.15
3 gall *
hyp...............
-1.548 1.125
-1.377 0.169
Records used 126
Deviance
80.8413 Free parameters
3
Informative strata 39 (handled with the conditional
likelihood)
Non-Informative strata 24
LRT
LR statistic
1.947 Degrees of
freedom 1
P value 0.163
We conclude this example with a categorical model for the
estrogen dose-response function. The model can be written as

where 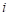 indexes the three conjugated
estrogen dose groups for exposed women (group 0 corresponds to unexposed women).
To fit this model, we first use the LEVELS command to
indicate that cdose is to be treated as a categorical variable. The model
is then fit in the usual way. The commands to carry out this analysis are
indexes the three conjugated
estrogen dose groups for exposed women (group 0 corresponds to unexposed women).
To fit this model, we first use the LEVELS command to
indicate that cdose is to be treated as a categorical variable. The model
is then fit in the usual way. The commands to carry out this analysis are
LEVELS cdose @
FIT cdose @
The output for this model is shown in Output
5.13.
Earlier we noted that information on conjugated estrogen dose
was not available for all women in the study and used the MISS command to recode the missing values to the EPICURE missing value
internal code. Only those case-control pairs in which both the case and her
matched control have nonmissing values for cdose should be used in the analysis.
If you look at the deviance summary in the figure, you will see that the number
of risk sets is given as 59, which is the number of case-control pairs for which
both women have known values for cdose. As a model is fit, PECAN, like all of the
EPICURE regression
programs, checks the variables involved in the fit for missing values and
automatically excludes those records with missing data. For PECAN, this also
involves checking whether a risk set can still be included in the analysis.
Additional topics related to missing values will be explored further in the next
example. The complete set of commands used in this example is shown in Listing 5.3 at the end
of this chapter.
Output 5.13 A categorical dose-response model - 1:1
Matching
LEVELS cdose @
cdose has 4 levels from 0 to 3
FIT cdose@
Iter Step Deviance
0 0 81.7914
1 0 63.5681
2 0 62.9863
3 0 62.9796
4 0 62.9796
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Stratification on setno with 59 strata
Conditional likelihood used for 59 strata
Using pairs == 1
cases is used for cases
Parameter Summary Table
#
Name
Estimate Std.Err. Test Stat. P value
-- ----------------------------
---------- --------- ---------- --------
Log-linear term 0
2
cdose_1..................
1.524 0.6181
2.466 0.0137
3
cdose_2..................
1.266 0.569
2.225 0.0261
4
cdose_3..................
2.120 0.693
3.059 0.00222
Records used
122 Unused
records 4
Deviance
62.9796 Free
parameters 3
Informative strata 45 (handled with the conditional
likelihood)
Non-Informative strata 14
Example
5.12 Matched Case-Control
Data with 1:M Matching
In the previous example, we considered a subset of the
endometrial study data in which the data were limited to the first matched
control for each case. In this example, we continue to work with the endometrial
cancer data, but we use all of the data, that is, four controls per case.
Because PECAN automatically detects 1:M or N:M (multiple cases and controls in
each stratum) matching and chooses the appropriate estimation algorithm, it is
not necessary for the user to do anything special when working with such data.
The commands to read the data are identical to those used in the previous
example and will not be repeated here. (One could carry out all of the analyses
described in this example in the same session as the analyses of the first
example by canceling the selection used to restrict the data to 1:1 matching
with the NOSELECT@ or SELECT@ command.)
We begin by fitting the null model, which assumes an odds ratio
of 1 in each matched set, and then we consider the model with a multiplicative
estrogen effect. The deviance for the null model is saved for use in the
computation of likelihood ratio tests and P-values. After computation of the
score statistic for an estrogen effect, we fit the estrogen effect model and
compute the likelihood ratio statistic (using the script file discussed above).
The commands to fit these models and carry out these tests are
FIT @ NULL
SCORE +est @
FIT @ LRT
The output from this analysis is shown below.
Output 5.14 Output from
fit of null model and estrogen effect model
FIT@
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Stratification on setno with 63 strata
Conditional likelihood used for 63 strata
cases is used for cases
Records used 315
Deviance
202.789 Free
parameters 0
Informative
strata 63 (handled with the conditional likelihood)
NULL
SCORE +est @
The score statistic is 31.1556 with df = 1 (P <
0.001 )
FIT @
Iter Step Deviance
0 0 202.789
1 0 168.806
2 0 167.462
3 0 167.443
4 0 167.443
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Stratification on setno with 63 strata
Conditional likelihood used for 63 strata
cases is used for cases
Parameter Summary Table
#
Name
Estimate Std.Err. Test Stat. P value
-- ----------------------------
---------- --------- ---------- --------
Log-linear term 0
1
est......................
2.074 0.4208
4.928 < 0.001
Records used 315
Deviance
167.443 Free
parameters 1
Informative strata 58 (handled with the conditional
likelihood)
Non-Informative strata 5
LRT
LR statistic
35.35 Degrees of
freedom 1
P value < 0.001
The format of the parameter summary table is identical to that
in the previous example. The point estimate of the log of the relative risk for
estrogen exposure, 2.074, is similar to that obtained in the restricted data set
of the previous example (2.269), but, because of the use of the additional data,
the standard error is about 30 percent less in the current analysis. Also, the
number of non-informative risk sets is much smaller in the full data set (5
sets) than it was in the 1:1 matched data (31 sets).
Thus far all of the PECAN examples have made use of the
classic linear model for the log of the relative risk. We will now consider an
alternative but, in this simple case, equivalent model in which the relative
risk is modeled as a linear function of estrogen usage. The model is

The commands to specify and fit this model are
NOMODEL
LINE 1 est @
FIT @
The first command resets the model, which in this case involves
removing the parameter associated with est from the log-linear subterm of
term 0, and is equivalent to a LOGL 0 @ command. If
the previous model contained parameters in several terms or subterms, the NOMODEL command provides a
convenient alternative to a series of empty subterm specification commands. The
model of interest is specified with the LINE 1 command
and fit with the FIT command. The parameter summary
table for this fit is shown below.
Output
5.15 Excess relative risk model with matched case-control data
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Stratification on setno with 63 strata
Conditional likelihood used for 63 strata
cases is used for cases
Parameter Summary Table
#
Name
Estimate Std.Err. Test Stat. P value
-- ----------------------------
---------- --------- ---------- --------
Linear term 1
1
est......................
6.955 3.348
2.078 0.0377
Records used 315
Deviance
167.443 Free
parameters 1
Informative strata 58 (handled with the conditional
likelihood)
Non-Informative strata 5
Because est is a binary
indicator variable, the deviance for this fit is identical to the previous
model. The parameter estimate, 6.955, is equal to 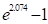 , where 2.074 is the estimate of the
log relative risk in the log-linear (multiplicative) risk model. Note that the
standard error for the excess relative risk parameter is larger than we would
expect based upon the P-value for the likelihood ratio test, P <0.001. (This
test is identical to that carried out for the log-linear model.) This is a
consequence of the parameterization (called parameter effects curvature in
modern, nonlinear regression literature) and indicates that one should not rely
on the Wald statistic
, where 2.074 is the estimate of the
log relative risk in the log-linear (multiplicative) risk model. Note that the
standard error for the excess relative risk parameter is larger than we would
expect based upon the P-value for the likelihood ratio test, P <0.001. (This
test is identical to that carried out for the log-linear model.) This is a
consequence of the parameterization (called parameter effects curvature in
modern, nonlinear regression literature) and indicates that one should not rely
on the Wald statistic  as a test for the statistical
significance of the estrogen effect in this model. It also indicates that
Wald-type confidence intervals for this parameterization are misleading. These
issues were discussed in detail in the GMBO/PECAN examples. One might obtain a
better asymptotic description of the variability in the excess relative risk by
estimating the logarithm of the excess relative risk using the model
as a test for the statistical
significance of the estrogen effect in this model. It also indicates that
Wald-type confidence intervals for this parameterization are misleading. These
issues were discussed in detail in the GMBO/PECAN examples. One might obtain a
better asymptotic description of the variability in the excess relative risk by
estimating the logarithm of the excess relative risk using the model
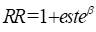
which could be specified and fit with the commands
LINE 1 est=1 @
LOGL 1 %CON @
FIT @
As discussed above and in the GMBO/PECAN examples, likelihood-based
confidence bounds provide a useful parameterization-independent description of
the variability in specific parameters (for equivalent models). The BOUNDS command can be used to compute likelihood-based
bounds for parameters in all EPICURE regression programs. The 95
percent likelihood bounds for the parameter in this linear model are (2.71,
18.78) while those for the log-linear model are (1.00, 2.93). You can easily
check the equivalence of these bounds.
Despite potential asymmetry in the likelihood surface,
alternative parameterizations such as those considered here are useful in some
problems. For example, if one is interested in dose-response models over a broad
range of exposures, it may be that the relative risk is better modeled as a
linear rather than log-linear function of dose. It may also be the case that
effects of factors that modify the risk are better modeled as effects on the
excess relative risk than as multiplicative effects on the relative risk.
We continue this example with an illustration of hypothesis
tests in the presence of missing values in some of the covariates of interest.
In particular, we will consider testing for a log-linear effect of conjugated
estrogen dose, cest, on the risk of endometrial cancer. After clearing
the previous linear model, one might use the commands
FIT @
FIT cest @
However, since conjugated estrogen dose is not available for
all women, the difference in the deviance for these two models is not a valid
likelihood ratio test for the estrogen dose response. The null model will be fit
using data on all women in the study (315 women in 63 matched sets), but the
dose response model will be based upon the data for the 291 women in 59 matched
sets with complete dose information. If the fact that estrogen dose is missing
is not related to subsequent cancer development, one can construct a valid
likelihood ratio test based on an analysis of the subset of women with dose
information. The CHECKMISS
command can be used to indicate one or more variables to be checked for missing
values prior to fitting each model. Records with missing values for any of the
variables in the check list are not included in subsequent analyses. Thus, we
can use the following commands to fit the null and full models in this
example:
CHECK cest @
FIT @
FIT cest @
The output produced by these commands is summarized in below.
Output
5.16 Likelihood ratio test with listwise deletion of records with
missing data
CHECK cest @
8 records rejected (in addition to the SELECT)
FIT @
Iter Step Deviance
0 0 159.223
1 0 159.223
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Using records with no missing values for: cest
Stratification on setno with 59 strata
Conditional likelihood used for 59 strata
cases is used for cases
Parameter Summary Table
#
Name
Estimate Std.Err. Test Stat. P value
-- ----------------------------
---------- --------- ---------- --------
Log-linear term 0
1
cest.....................
1.710 0.3541
4.83 < 0.001
Records used
307 Unused
records 8
Deviance
159.223 Free
parameters 1
Informative strata 56 (handled with the conditional
likelihood)
Non-Informative
strata 3
FIT cest @
Iter Step Deviance
0 0 188.129
1 0 159.498
2 0 159.224
3 0 159.223
4
0 159.223
Conditional-logistic regression
Additive excess relative risk T0 * (1 + T1 + T2 +
...)
Using records with no missing values for: cest
Stratification on setno with 59 strata
Conditional likelihood used for 59 strata
cases is used for cases
Parameter Summary Table
#
Name
Estimate Std.Err. Test Stat. P value
-- ----------------------------
---------- --------- ---------- --------
Log-linear term 0
1
cest.....................
1.710 0.3541
4.83 < 0.001
Records used
307 Unused
records 8
Deviance
159.223 Free
parameters 1
Informative
strata 56 (handled with the conditional likelihood)
Non-Informative strata 3
Based on the large change in the deviance, it is clear that
there is a dose response, with the risk changing by a factor of 5.5 per unit of
coded estrogen dose.
The commands for this example are shown in Listing 5.4 at the end of the chapter.
 . Because of the way in which the data
are sorted, the value of caseno is 1 for the cases and 2 for the first
control for each case. The pairs variable is an indicator used to select records
for the initial analyses. The data are saved in a BSF file with the default
name, LEIMOD.BSF.
. Because of the way in which the data
are sorted, the value of caseno is 1 for the cases and 2 for the first
control for each case. The pairs variable is an indicator used to select records
for the initial analyses. The data are saved in a BSF file with the default
name, LEIMOD.BSF.